Hello evoryone,
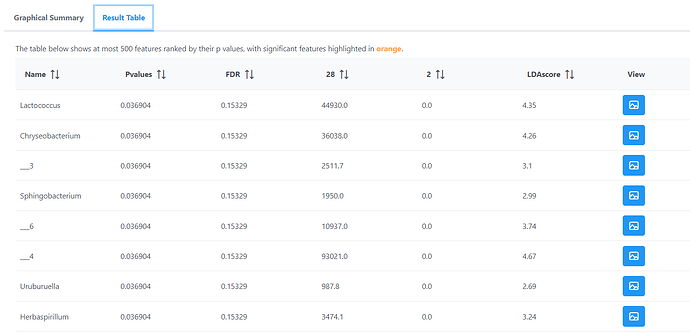
I’m using Microbiome Analyst weebtool to analyse data preously processed with Qiime2. I have experience with this weebtool, and I know that normally works well with my data. Only when using data collapsed at genus level, when I try differential analyses like LEfSe at Feature-level, tags like “__3” start to appear, and it’s imposible to track what feature really is.
I upload some data where the problem is present. yo can also see the tag problem in the screenshot.
Thank you for your help.
Best wishes.
OTUs-6collapse.csv (215.6 KB)
Metadata.csv (106 Bytes)