Hi,
I have 132 sequence files (234 if R1/R2). I used DADA2 pipeline of NEPHELE to create ASV with biom, taxanomy table and tree. I tried to use DADA2 pipeline of qiime 2 but there were additional error hence I switcehed to Nephele. Here are some of the errors I am facing
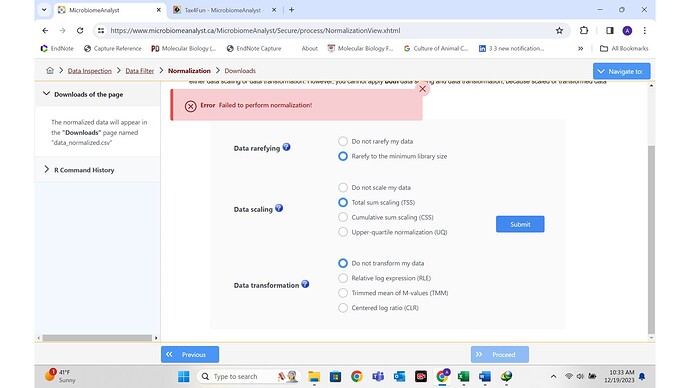
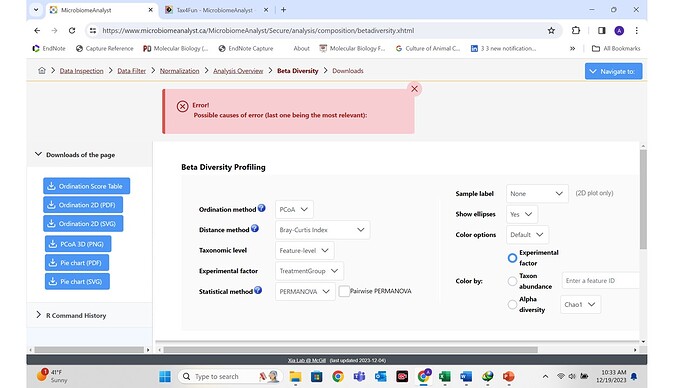
Unfortunately my files cannot be rarifed to minimum library size and I cannot generate beta diversity. Instead an error pops without a explanation.
I have used Microbiomenalayst for a publication in past. This is the first time its happening. I am ready to upload my files.
Taxonomy table_121823_excel edited.txt (312.9 KB)
Non-rarefied ASV table_121823_excel edited.txt (1.3 MB)
Taxonomy_table_121823.txt (389.2 KB)
Non-rarefied ASV table_121823.txt (1.5 MB)
N2_mappingfile_2023_2.csv (24.9 KB)