-
NetworkAnalyst currently accepts list(s) of genes or proteins: one or more lists of gene or protein IDs ( from 17 species) with optional expression profiles (i.e. fold changes, p values). Each gene should be in a row. Please refer to our example data for a single list, or multiple lists.
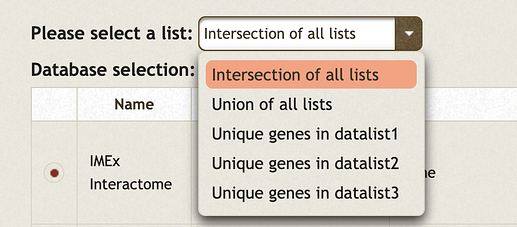
- Please note when multiple gene/protein lists are uploaded, users will be asked to choose among possible combinations among them for network creation. See an screenshot below (3 lists uploaded)
- Please note when multiple gene/protein lists are uploaded, users will be asked to choose among possible combinations among them for network creation. See an screenshot below (3 lists uploaded)
-
Network file (any species): users can upload network files generated with a different software to perform network visualization in NetworkAnalyst. Four different formats are supported including .sif, .json, .txt and .graphml format. Please click on the following links to see example files supported:
NetworkAnalyst is mainly designed for gene/protein lists. We also have three tools for network creation for other purposes:
- OmicsNet: creating general purpose multi-omics network linking all common molecules (including LC-MS peaks and microbiome data!)
- miRNet: creating miRNA-centric network linking miRNAs with genes, TFs, SNPs, etc
- mGWAS-explorer: connecting metabolites, genes, SNPs