OmicsNet builds interaction networks starting from list(s) of input molecules: gene/protein, TF, miRNA, metabolite, and supports the integration of multiple interaction types. OmicsNet can also process and integrate list of microbial taxa, peaks generated from untargeted metabolomics and SNPs to the molecular interaction framework. The workflow can be dissected in four steps:
- Upload one or more list(s) of molecules.
- Select interaction database and compute individual omics interaction networks for each input list(s). Optionally, further expanding current network - if there is no shared nodes or direct connections to allow network merging (next step);
- Merge above individual networks into connected subnetworks. Optionally, perform network trimming and filtering - if networks are too big;
- Explore networks in 2D or 3D space.
The main goal of OmicsNet is to allow easy creation of knowledge-based networks from different input lists of molecules. Here are some common scenarios:
Scenario 1: upload a list of omics features and search against different interaction databases using the seed features from list upload as query. An example of network construction starting from gene list is shown below:
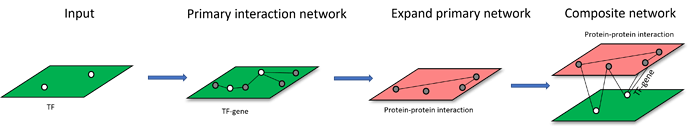
Scenario 2: upload a list of omics features, build a primary interaction network and expand the primary interaction networks by adding other types of interactions by using nodes from primary interaction network as query. An example of network construction starting from TF list is shown below:
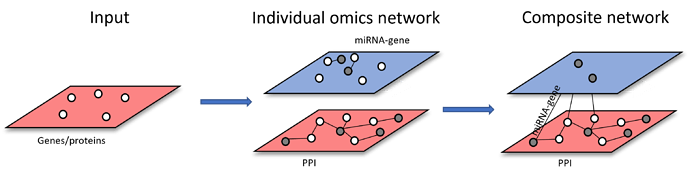
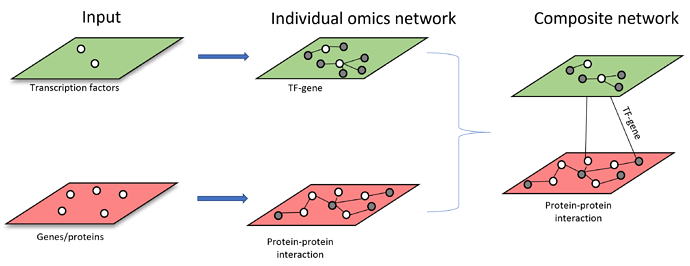
Scenario 3: starting from more than one types of omics features. Each list of omics features will be used to build individual omics interaction networks. The resulting individual networks will be merged through shared nodes to form multi-omics network (i.e. Metabolite-protein and metabolite-taxa networks are merged through shared metabolites across both networks)
The above approaches will typically return one giant subnetwork (“continent”) with multiple smaller ones (“islands”). Most subsequent analyses are performed on the continent. Note,
Networks with less than 3 nodes will be excluded.
We will be happy to hear if you have invented new use cases of OmicsNet!